Abstract
Introduction: A risk-adapted treatment strategy is of crucial importance in patients with myelodysplastic syndromes (MDS). With the rapid development of high through-put technology like next generation sequence (NGS), multiple mutations have been revealed as significant factors in MDS. However, previous risk prognostic scoring systems such as the International Prognostic Scoring System (IPSS), the revised IPSS (IPSS-R), and the World Health Organization-classification-based Prognostic Scoring System (WPSS) did not integrate molecular abnormalities. The newly published IPSS-Molecular (IPSS-M) model, which combining genomic profiling with hematologic and cytogenetic parameters in the IPSS-R system (including bone marrow blasts, platelets and hemoglobin), was recently developed to evaluate the associations with leukemia-free survival (LFS), leukemic transformation, and overall survival (OS). This model is considered to be a valuable tool for clinical decision-making. However, it has not yet been widely validated in the clinic. This study is to further validate the prognostic power of IPSS-M based on the real-world data, and to compare the prognostic value of different scoring systems in patients with MDS.
Methods:A population of 255 adult MDS was considered. The IPSS-M Web calculator (https://mds-risk-model.com) was used to calculator a tailored IPSS-M score of each patient, and the risk category was therefore divided correspondingly. We then compared the IPSS-M prognostic power to that of IPSS, IPSS-R, and WPSS. The receiver operating characteristic curve (ROC) was used to evaluate the efficiency of the prognostic models. The Spearman's rank correlation rho was used to evaluate the relevance of different tools. Kaplan-Meier (K-M) survival curve with Log-Rank analysis was performed in the study.
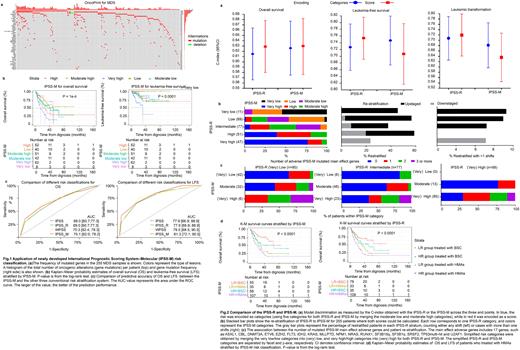
Results: Median follow-up was 33.2 (95%CI: 27.8-41.1, reverse K-M) months for the whole cohort. Median OS was 28.3 (21.3-35, K-M) months. We mapped at least one oncogenic gene alteration in 75% of patients with MDS, and found that TET2 (23%), ASXL1 (19%), TP53 (15%), DNMT3A (10%), U2AF1 (9%), SF3B1(9%), EZH2 (9%), RUNX1 (5%), WT1 (5%) and SETBP1(5%) are the top 10 genes with the highest mutation frequency (Fig. 1a). Similar with the other three prognostic systems, IPSS-M risk classification was statistically significant for OS (P<0.0001 for IPSS-M, P=0.00061 for IPSS, P<0.0001 for IPSS-R and WPSS) and LFS (P<0.0001 for IPSS-M, IPSS, IPSS-R and WPSS) (Fig 1b). However, no prognostic tool predicted the probability of achieving an objective response. Among the four risk stratification tools, WPSS was more superior in sensitivity and accuracy for OS, while IPSS-M was more superior for LFS (Fig. 1c). The Spearman's rho of IPSS-M with IPSS, IPSS-R and WPSS was 0.646, 0.786 and 0.729, respectively. This indicated that IPSS-M was more correlated with IPSS-R. The mapping C-index between IPSS-R and IPSS-M categories resulted in improved discrimination across the OS and LFS but not leukemic transformation (Fig. 2a). Additionally, it is also resulted in the re-stratification of 45% of MDS patients (114/255) (Fig. 2b). Of these, 85 (33%) were upstaged and 29 (11%) were down-staged. Among re-stratified patients, 19 of 80 (24%) had one mutated gene in IPSS-M, whereas 23 of 80 (29%) had two or more (Fig. 2c). Thus, patient re-stratification was not a single gene effect, but the cumulative contribution of the prognostic mutations for each patient. To further explore the effect of hypomethylation agents (HMAs) or best supportive care (BSC) on different groups, we analyzed the K-M survival curves of higher-risk (HR) and lower-risk (LR) groups stratified by IPSS-M and IPSS-R (Fig. 2d). The results showed that compared with BSC, HMAs did not improve OS and LFS either in HR or LR groups. In patients treated with HMAs, IPSS-M was more superior in both sensitivity and applicability than IPSS-R in OS, with the C-index value of 0.611 vs. 0.582.
Conclusions: Overall all prognostic scoring systems well represent the prognostic risk of MDS patients. A highly significant correlation was found between the IPSS-M and IPSS-R risk classifications. IPSS-M provides relevant information for the implementation of risk-adapted strategies in MDS.
Disclosures
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal